Scientific software can be confusing, particularly when you're doing something that the software isn't primarily intended for. I often find myself wanting to run quick-and-dirty molecular dynamics simulations on small organic molecules, but I've struggled to find an easy way to do this using open-source tools like OpenMM.
This is particularly frustrating since I feel like I should be equipped to succeed at this task:
Yet despite all this, I've probably tried and failed to use OpenMM for my research a half-dozen times over the past five years (evidence). I always get bogged down somewhere: I don't have PDB files for my compounds, or I can't figure out how to get the forcefield parameters right for a small molecule, or I just get lost in a sea of similar-sounding classes and objects. Part of the problem here is that all the "intro to MD" tutorials seem to assume that you're hoping to run an MD simulation on a protein from the PDB—so if you have an .xyz file, it's not obvious how to proceed.
Nevertheless, I've finally succeeded. Here's the code, with minor annotations interspersed:
from openff.toolkit import Molecule, Topology from openmm import * from openmm.app import * import nglview import mdtraj import matplotlib.pyplot as plt import numpy as np import openmoltools import tempfile import cctk import openmmtools import math from random import random, randint from sys import stdout import pandas as pd from rdkit import Chem from rdkit.Chem import AllChem from openmmforcefields.generators import SMIRNOFFTemplateGenerator %config InlineBackend.figure_format='retina'
Already, we're off to a complex start: we need OpenFF, OpenMM, openmoltools, openmmtools, and openmmforcefields (not to mention nglview and mdtraj). There's a broader point to be made here about the state of scientific software and how this relates to academic incentive structure, but I digress...
smiles = "c1cc(F)ccc1O"
def generate_forcefield(smiles: str) -> ForceField:
""" Creates an OpenMM ForceField object that knows how to handle a given SMILES string """
molecule = Molecule.from_smiles(smiles)
smirnoff = SMIRNOFFTemplateGenerator(molecules=molecule)
forcefield = ForceField(
'amber/protein.ff14SB.xml',
'amber/tip3p_standard.xml',
'amber/tip3p_HFE_multivalent.xml'
)
forcefield.registerTemplateGenerator(smirnoff.generator)
return forcefield
def generate_initial_pdb(
smiles: str,
min_side_length: int = 25, # Å
solvent_smiles = "O",
) -> PDBFile:
""" Creates a PDB file for a solvated molecule, starting from two SMILES strings. """
# do some math to figure how big the box needs to be
solute = cctk.Molecule.new_from_smiles(smiles)
solute_volume = solute.volume(qhull=True)
solvent = cctk.Molecule.new_from_smiles(solvent_smiles)
solvent_volume = solvent.volume(qhull=False)
total_volume = 50 * solute_volume # seems safe?
min_allowed_volume = min_side_length ** 3
total_volume = max(min_allowed_volume, total_volume)
total_solvent_volume = total_volume - solute_volume
n_solvent = int(total_solvent_volume // solvent_volume)
box_size = total_volume ** (1/3)
# build pdb
with tempfile.TemporaryDirectory() as tempdir:
solute_fname = f"{tempdir}/solute.pdb"
solvent_fname = f"{tempdir}/solvent.pdb"
system_fname = f"system.pdb"
smiles_to_pdb(smiles, solute_fname)
smiles_to_pdb(solvent_smiles, solvent_fname)
traj_packmol = openmoltools.packmol.pack_box(
[solute_fname, solvent_fname],
[1, n_solvent],
box_size=box_size
)
traj_packmol.save_pdb(system_fname)
return PDBFile(system_fname)
def smiles_to_pdb(smiles: str, filename: str) -> None:
""" Turns a SMILES string into a PDB file (written to current working directory). """
m = Chem.MolFromSmiles(smiles)
mh = Chem.AddHs(m)
AllChem.EmbedMolecule(mh)
Chem.MolToPDBFile(mh, filename)
forcefield = generate_forcefield(smiles)
pdb = generate_initial_pdb(smiles, solvent_smiles="O")
system = forcefield.createSystem(
pdb.topology,
nonbondedMethod=PME,
nonbondedCutoff=1*unit.nanometer,
)
This code turns a SMILES string representing our molecule into an OpenMM System, which is a core object in the OpenMM ecosystem. To do this, we have to do a lot of shenanigans involving figuring out how many solvent molecules to add, calling PACKMOL, etc. One of the key steps here is the SMIRNOFFTemplateGenerator (documented here), which uses one of the recent OpenFF forcefields to describe our chosen molecule.
# initialize Langevin integrator and minimize
integrator = LangevinIntegrator(300 * unit.kelvin, 1 / unit.picosecond, 1 * unit.femtoseconds)
simulation = Simulation(pdb.topology, system, integrator)
simulation.context.setPositions(pdb.positions)
simulation.minimizeEnergy()
# we'll make this an NPT simulation now
system.addForce(MonteCarloBarostat(1*unit.bar, 300*unit.kelvin))
simulation.context.reinitialize(preserveState=True)
checkpoint_interval = 100
printout_interval = 10000
# set the reporters collecting the MD output.
simulation.reporters = []
simulation.reporters.append(DCDReporter("traj_01.dcd", checkpoint_interval))
simulation.reporters.append(
StateDataReporter(
stdout,
printout_interval,
step=True,
temperature=True,
elapsedTime=True,
volume=True,
density=True
)
)
simulation.reporters.append(
StateDataReporter(
"scalars_01.csv",
checkpoint_interval,
time=True,
potentialEnergy=True,
totalEnergy=True,
temperature=True,
volume=True,
density=True,
)
)
# actually run the MD
simulation.step(500000) # this is the number of steps, you may want fewer to test quickly
Here, we configure the settings that will be familiar to people who read MD textbooks. I've chosen to use a Langevin integrator/thermostat to control temperature, and added a Monte Carlo barostat to make this an NPT simulation (constant pressure & temperature). This isn't appropriate for all uses, but it means we don't have to worry about getting the box size exactly right. We also configure how we're going to save data from the simulation, and then the last line actually runs it (this might take a while if you don't have a GPU).
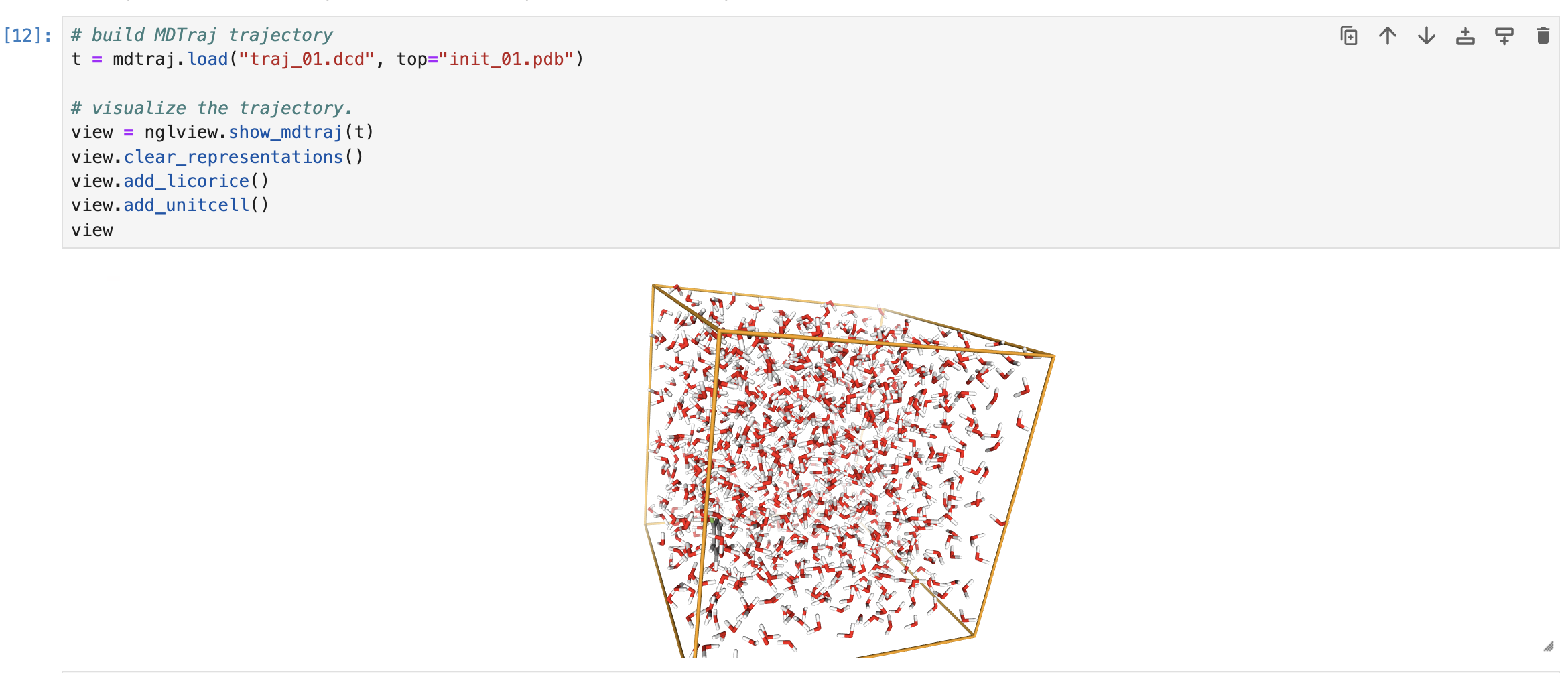
Once we've run this simulation, we can visualize the output and watch things wiggle around! The MDTraj trajectory is a pretty versatile object so you can do much more analysis here if you want to.
# build MDTraj trajectory
t = mdtraj.load("traj_01.dcd", top="init_01.pdb")
# visualize the trajectory - this works in jupyter at least
view = nglview.show_mdtraj(t)
view.clear_representations()
view.add_licorice()
view.add_unitcell()
view

It's also good to check that the thermostat actually worked and nothing blew up:
# check that the thermostat is working
df = pd.read_csv("scalars_01.csv")
df.plot(kind="line", x='#"Time (ps)"', y="Temperature (K)")
This is probably laughably simple to an OpenMM expert, but it works well enough for me. Here's my current list of things I still don't know how to do:
Still, I hope this script is a useful asset to the community, and helps other people not make the same mistakes I did.
Thanks to Dominic Rufa for answering some of my stupid questions.